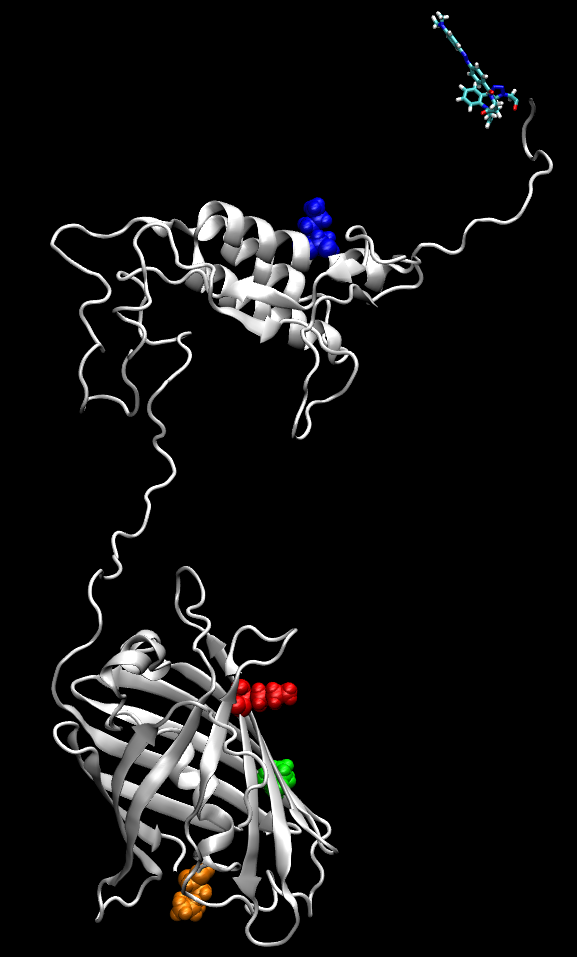
We develop strategies based on a combined in silico/in vitro adaptive approach to generate molecular constructs to be used as diagnostic tests, using minimal parts of the pathogen’s proteins and its molecular targets, and with even partial knowledge of the interaction interface. The detection method is based on fluorescence techniques capable of identifying the interaction between the viral element and the specific cellular receptor by exploiting the phenomena of Fluorescence Resonance Energy Transfer (FRET) or quenching between pairs of fluorophores. The detection is immediate, does not require post-processing, and can be carried out at low cost with a simple fluorimetric device.
The innovation potential of this research line consists in its flexibility: the current techniques of protein expression through both prokaryotic organisms, such as E. coli and eukaryotic, such as yeasts, allow to produce protein constructs with high efficiency and specificity given the sequence and then test their efficacy directly in vitro. In the case of appearance of new virus mutations, the modeling techniques allow to change the sequences in a virtual way and to estimate their affinity to the target before the experimental tests in order to promptly reorient and optimize the subsequent development of the new construct detector.
| People | Antonella Battisti, Ranieri Bizzarri, Giorgia Brancolini*, Daniele Montepietra, Riccardo Nifosì, Antonella Sgarbossa, Barbara Storti, Valentina Tozzini |
| Keywords | FRET biosensors; simulation of biological systems; biomedical engineering; enzymatic assays; SARS-Cov-2 COVID |
| Methods, techniques | Molecular Dynamics; Force Field Development; calculation of FRET efficiency; Coarse Grained Models |
| Granted projects | |
| 2022-2025: PRIN2020 Project (cod. 2020LW7XWH) “Early Phase Preclinical Development of PACECOR, a Mutation-Independent Anti-SARS-Co2 Therapeutic Strategy” (PI G. Brancolini) | |
| 2020-2022: Supercomputing project “Molecular strategies to inhibit the COVID-19-cell interaction” at ORNL, USA. CNMS2020-B-00433. Granted 1.000.000 CPU hours (2 years). (PI G. Brancolini) | |
| Collaborations | Eleonora Da Pozzo, Laura Marchetti, UniPI Dept Pharmaceutical Technologies |