Molecular condensates, i.e. membraneless dynamic nanostructures are ubiquitous in cell’s life because they enable control over complex biochemical reactions in space and time. High/super-resolution fluorescence microscopy is perfectly suited for the study of molecular condensates, because it leverages sensitivity down to single-molecule, color-coded molecular information, and ability to visualize interactions and structures at nanoscale. In this context, we focus on two general kinds of condensates:
1. Molecular condensates that regulate gene expression. Organization of the genome into the transcriptionally active euchromatin and silenced heterochromatin is essential for eukariotic cell function. Polycomb complexes (PRC1 and PRC2) yield condensates able to modify translationally histones, thereby generating transient nanoscale domains of silenced (“facultative”) heterochromatin which control transcription and regulate gene expression. Loss of this competence is crucially linked to several tumors. We focus on PRC1/2 dynamic assembly onto chromatin and pharmacological strategies to address them.
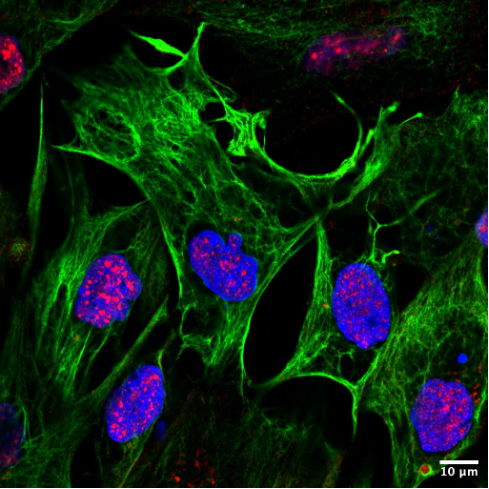
2. Lipid-raft organization of plasma membrane receptors. Lipid-rafts are dynamically fluctuating nanoscale assemblies of sphingolipid, cholesterol, and proteins on the cell plasma membrane that can be stabilized to coalesce, forming platforms that function in membrane signaling and trafficking. We investigate membrane protein receptors such as ACE2 (the receptor of SARS-CoV-2) and PD-L1 (a crucial molecular determinant of immunotolerance harnessed by tumor machinery for evasion, and one of the targets of current immunotherapies in many cancers) whose subtle interplay with lipid rafts is still obscure and meant to play a fundamental role in their biological activities.
For these goals, we engineer novel fluorescent proteic and organic probes and imaging schemes able to report on the biophysical properties of cellular condensates at nanoscale.
| People | Ranieri Bizzarri, Vincenzo Canestrale, Barbara Storti* |
| Keywords | Molecular condensates, protein aggregation, polycomb proteins, lipid rafts, ACE2, PD-L1 |
| Methods, techniques | ISM, Image Scanning Microscopy; SMLM, Single Molecule Localization Microscopy; STED, STimulated Emission Depletion; STORM, STochastic Optical Reconstruction Microscopy; TIRF, Total Internal Reflection Fluorescence; cell cultures; immunostaining |
| Granted projects | 2020-2022 Università di Pisa PRA 2020_2021 “Indagare gli effetti di interferenti endocrini sulla funzione e cancerogenesi tiroidea“ |
| Collaborations | Prof. Vittoria Raffa, Department of Biology, University of Pisa (IT), Prof. Romano Danesi, Department of Clinical Medicine, University of Pisa (IT) and Pisa University Hospital, Dr. Alessandra Montecucco, Institute of Molcular Genetics-CNR (IGM-CNR), Pavia, Italy, Gianmarco Ferri, Fondazione Pisana per la Scienza, Pisa (IT), Alberto Diaspro, Nanoscopy, CHT, Istituto Italiano di Tecnologia, Genoa (IT) |
| Publications | |
| G Maroni, MA Bassal, I Krishnan, CW Fhu, V Savova, R Zilionis, VA Maymi, N Pandell, E Csizmadia, J Zhang, B Storti, J Castaño, P Panella, J Li, CE Gustafson, S, Fox, RD Levy, CV Meyerovitz, PJ Tramontozzi, K Vermilya, A De Rienzo, S Crucitta, DS Bassères, M Weetall, A Branstrom, A Giorgetti, C Ciampi, M Del Re, R, Danesi, R Bizzarri, H Yang, O Kocher, AM Klein, RS Welner, R, Bueno, MC Magli, JG Clohessy, DG Tenen, E Levantini, Identification of a targetable KRAS-mutant epithelial population in non-small cell lung cancer Comm. Biol. 2021 | |
| S De Vincentiis, A Falconieri, M Mainardi, V Cappello, V Scribano, R Bizzarri, B Storti, L Dente, M Costa, V Raffa, Extremely Low Forces Induce Extreme Axon Growth J Neuroscience 2020 | |
| B Storti, S Civita, P, Faraci, G Maroni, I Krishnan, E Levantini, R Bizzarri Fluorescence imaging of biochemical relationship between ubiquitinated histone 2A and Polycomb complex protein BMI1 Biophys Chem 2019 | |
| G Abbandonato, B Storti, I Tonazzini, M Stöckl, V Subramaniam, C Montis, R Nifosì, M Cecchini, G Signore, R Bizzarri Lipid-conjugated rigidochromic probe discloses membrane alteration in model cells of Krabbe disease Biophys. J. 2019 | |
| B Storti, E Margheritis, G Abbandonato, G Domenichini, J Dreier, I Testa, G Garau, R Nifosì, R Bizzarri, Role of Gln222 in Photoswitching of Aequorea Fluorescent Proteins: A Twisting and H-Bonding Affair? ACS Chem. Biol. 2018 |